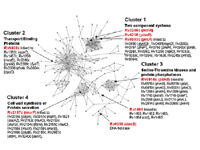
Network Analysis of Humans, Pathogens and Environmental Factors

Human disease often results from a complex interplay among human genetic factors, human-pathogen interactions, and environmental stimuli. It is therefore one of the goals of our group to better understand the mechanisms of human disease by investigating the relationships among these factors through the generation and analysis of host-pathogen-environmental networks. These networks take into account the genomic background of an individual and associated pathogens, as well as cellular interactions including protein-protein interactions, gene-disease relationships, chemical-gene relationships, chemical-disease relationships, and other. Together we hope that this type of network analysis will shed light on the molecular mediators of human disease, and provide clues to better understand and combat various diseases.
Expression Profiling of Humans and Associated Pathogens
In collaboration with Nick Walter (National Jewish Health) and Charles Daley, MD, (National Jewish Health), we will be examining the gene expression profiles of humans with varying degrees of tuberculosis disease and other respiratory diseases. We will also be examining the gene expression profiles of M. tuberculosis isolated from these same individuals with the goal of identifying host and pathogen gene signatures of disease, as well as to identify important pathways involved in disease, disease progression, and disease response.
Genome Sequencing, Analysis and Structural Bioinformatics
Together with Morten Sommer, PhD, (Technical University of Denmark) we will be sequencing the complete genomes of select M. tuberculosis isolates from the National Jewish Health repository. Particularly those that are resistant to first and second line antibiotics. We will combine the characterization and analysis at the phenotypic and genotypic levels to identify novel mechanisms of drug resistance, as well as to aid in the determination of more effective therapeutic regiments.
We also have an ongoing collaboration with Megan Murray, MD, DPH (Opens in a new window) (Opens in a new window) (Harvard School of Public Health) and members of the Broad Institute, examining the mechanisms of drug resistance in MDR and XDR M. tuberculosis isolates using a combination of genome sequencing, structural modeling and analysis, and literature mining. This strategy has important implications ranging from the design and discovery of novel inhibitors to the design of comprehensive gene-based diagnostics.
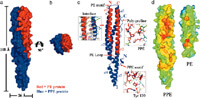
We are also very interested in utilizing the tools of structural biology and structural informatics to garner a better understanding of the impact of genomic polymorphisms among humans and human pathogens.