Current Projects
Cytokine Biology
We have a long-standing interest in studying cytokine biology. We investigated the roles of STAT1, STAT5 and STAT6 in the differentiation of Th1, Th2, eosinophils, basophils and mast cells. Super-enhancers comprise larger tracts of genomic DNA that include multiple smaller constituent enhancers. Super-enhancers are often associated with genes that confer cell identities and are enriched in genetic variants that may contribute to disease progression and severity. These enhancers contain a variety of consensus transcription factor binding motifs that are capable of binding to transcription factors that are activated by various external stimuli (Figure 1). We found that many cytokine and chemokine genes are associated with super-enhancers. Currently, we are investigating how super-enhancers at the cytokine and chemokine genes and their associated transcription factors integrate signals triggered by various external stimuli, such as antigenic stimulation, infection, cytokine and metabolic products, to generate hyper transcriptional outputs. We use monomethylation of lysine residue four on histone 3 (H3K4me1) and acetylation of lysine residue 27 on histone 3 (H3K27ac) modifications to identify the potential enhancers and use Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq) to identify transcription factor binding motifs within enhancers. We determine the function of enhancers and their associated transcription factors in primary cells, cell line and mice using the CRISPR-mediated transcription factor binding site editing or transcriptional repression. We study super-enhancers at cytokine and chemokine genes in effector cells, including helper T cells, innate lymphoid cells, basophils and mast cells, lung epithelial cells and macrophages. We determine how the super-enhancers detect transcriptional signals triggered by external stimuli and convert the signals into transcriptional outputs in the diseased immune conditions, such as virus infection, food allergy, asthma and autoimmune disease.
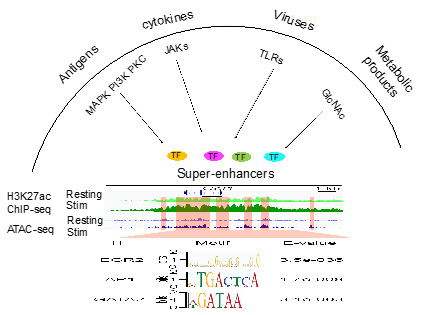
Figure 1. Super-enhancers contain a variety of consensus transcription factor binding motifs that bind to transcription factors that are activated by various external stimuli.
Regulation of the Genes that Control Immune Effector Differentiation
One of the important questions in immunology is how immune effector cells acquire the ability to express a set of genes whose products confer them with the specialized immune functions. We investigate how master transcription factors regulate the set of genes during the differentiation of progenitors/precursors into immune effector cells. As illustrated in Figure 2, master transcription factors GATA2 and MITF act as pioneering factors to “Open Up” chromatin of their targeted transcription factor genes, such as mast cell-specific transcription factors MITF, mast cell-specific transcription factor 1 (MCTF1) and MCTF2, then act together with MITF, MCTF1 and MCTF2 along with common transcription factors, such as SP1 and AP1, to regulate their target genes whose products carry out immune functions in a feed-forward manner.
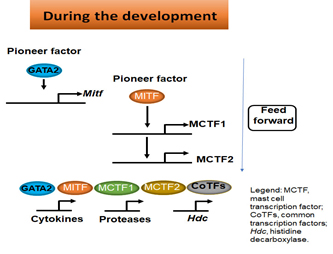
Figure 2. Pioneer factors induce a network of transcription factors that control the expression of genes that confer effector cells with specified functions.
Contribution of Genetic Factors to Enhancer Activity
Because some individuals are more susceptible to develop certain diseases and develop more severe forms of the diseases, we are also interested in studying how regulatory variants found in the promoters and enhancers contribute to disease susceptibility and severity. Single nucleotide differences found in transcription factor binding motifs within the promoters and enhancers can create or disrupt the binding of transcription factors to their target enhancers. These polymorphisms in transcription factor binding sites are most likely to influence the expression of the genes that play critical roles in disease development and severity. We are interested in identifying and validating these polymorphisms using various genome-wide enhancer enrichment technology and bioinformatics tools. We determine whether these polymorphisms affect the recruitment of transcription factor complexes using transcription factor binding-specific proteomics.
