Colorado CF RDP Molecular and Genomics Core
The ability to identify, analyze, and compare isolates within the same subspecies of M. abscessus or MAC is central to understanding the diversity of infectious disease pathogens and identifying the occurrence of possible patient-to-patient transmission within CF Centers. Analysis is also needed to identify the possibility of a shared source of environmental exposure, or to rule-out such an environmental source. Thus, genomic sequencing and phylogenomic analysis of isolates is essential to identify outbreaks and limit their occurrence and spread.
The Molecular Core accepts samples from the NTM Culture, Biorepository, and Coordinating Core. Samples are analyzed using whole genome sequencing (WGS) and comparative phylogenomic methods. These results are communicated back to the NTM Culture, Biorepository, and Coordinating Core, for dissemination to the referring Centers. The primary goal of the Molecular Core is to characterize the diversity and prevalence of NTM species affecting the CF community, and to accurately identify and limit potential instances of patient-to-patient transmission or nosocomial acquisition events in CF Centers. Click here to submit a CF patient NTM sample.
This work is the culmination of contributions from members of the Colorado Cystic Fibrosis Research Develpment Program (CF RDP) and collaborators, including Nabeeh A. Hasan, Rebecca M. Davidson, L. Elaine Epperson, Sara Kammlade, Rachael Rodger, Adrah Levin, Alyssa Sherwood, Scott Sagel, Stacey Martiniano. Charles L. Daley, Max Salfinger, Jerry A. Nick, and Michael Strong.
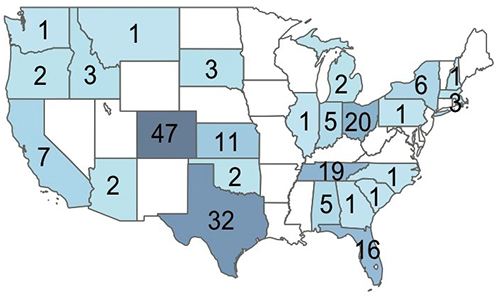
The Colorado Cystic Fibrosis RDP has sequenced hundreds of NTM isolates from CF patients across the United States.
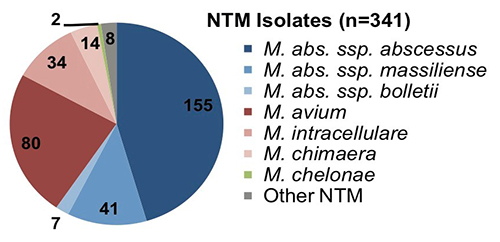
A summary analysis of the first 341 NTM isolates is available in our BioRxiv preprint:
Population Genomics of Nontuberculous Mycobacteria Recovered from United States Cystic Fibrosis Patients.
Nabeeh A. Hasan*, Rebecca M. Davidson*, L. Elaine Epperson, Sara M. Kammlade, Rachael R. Rodger, Adrah R. Levin, Alyssa Sherwood, Scott D. Sagel, Stacey L. Martiniano. Charles L. Daley, Max Salfinger, Jerry A. Nick, Michael Strong. BioRxiv DOI 10.1101/663559 (2019)
First 341 CF NTM Isolates Sequenced, Shown by Adjusted State of Origin.
Of the first 341 sequenced NTM isolates, 60% are Mycobacterium abscessus species and 38% are Mycobacterium avium complex